paired end sequencing vs mate pair
I would assume if it is not specified a library is a paired-end. There is a unique adapter sequence on both ends of the paired-end read labeled Read 1 Adapter and Read 2 Adapter.
De Novo Genome Assembly Using Next Generation Sequence
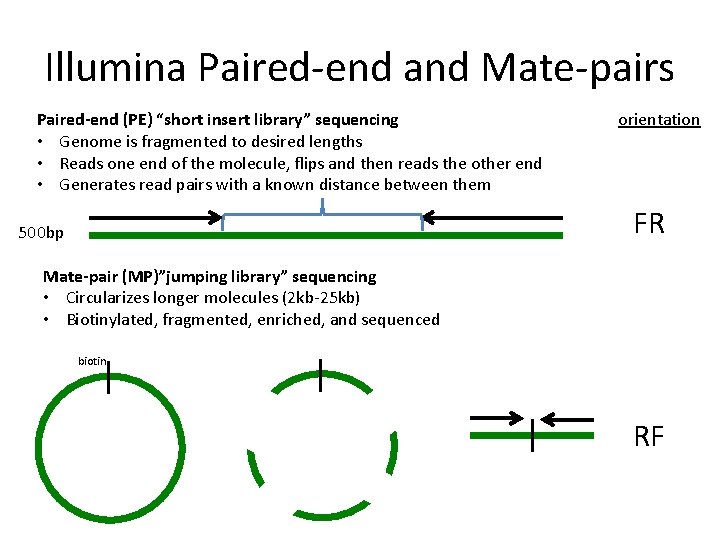
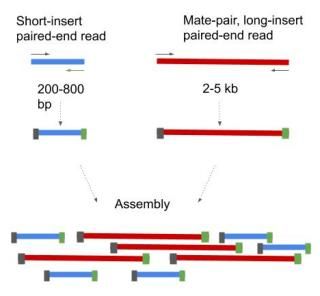
Using a combination of short and long insert sizes with paired-end sequencing results in maximal coverage.

. Reads come in pairs. Paired end sequencing vs Mate pair library sequencing Sanger chemistry를 이용한 전통적인 샷건 시퀀싱에 익숙해 있는 내게 paired end sequencing이나 mate pair library sequencing이나 다를 바가 없다. Some specialized technologies such as using circularized DNA fragments to create large insert jumping libraries Talkowski 2011 switch things around so that your reads ought to.
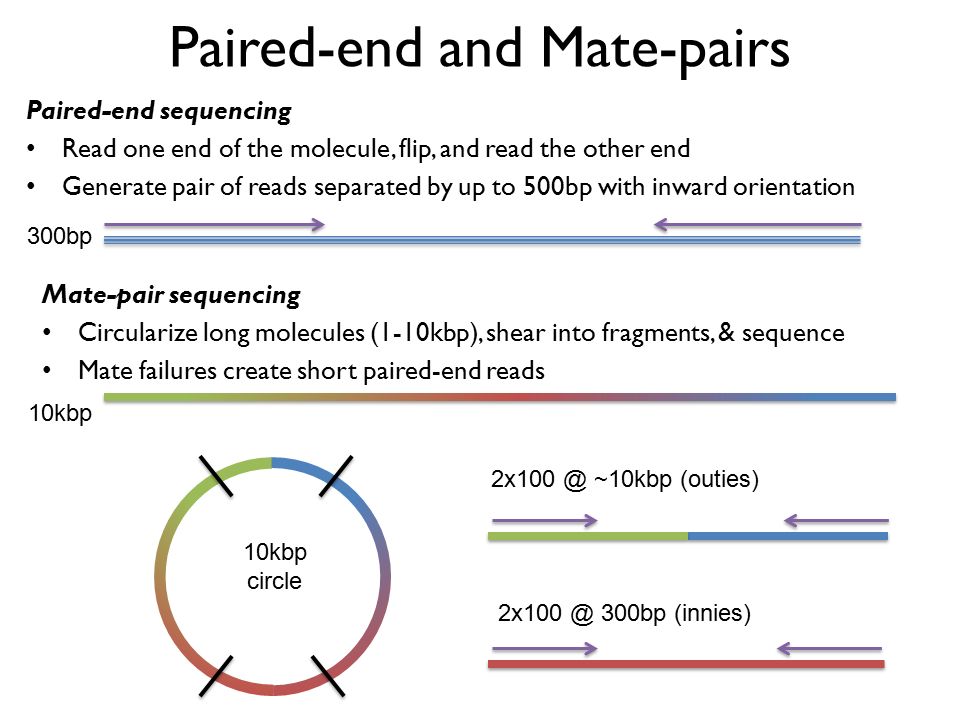
In paired-end sequencing the library preparation yields a set of fragments and the machine sequences each fragment from both ends. In contrast mate pairs arise from a fragment that is circularized before sequencing. The similarities between PE and MP reads include.
Ad Gene Expression Profiling Chromosome Counting Epigenetic Changes Molecular Analysis. Methods A novel algorithm that utilizes a suffix array has been specifically designed to compute the uniqueness of paired reads with fixed or variable mate-pair distance. In mate-pair sequencing the library preparation yields two.
That means that R1 is oriented forward. In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment. 그런데 며칠 전 Illumina의 시퀀싱에 대해 검토하다가 이 두 가지 방법을 다르게 기술하고.
We obtain theoretical read length lower bounds for re-sequencing that are also applicable to paired-end de novo assembly. Scaffolds NNN Contigs Illumina Reads Assembly Strategy Paired-end reads Mate-pair reads NNNN Contiging Scaffolding. First PE paired end reads are typically short 50-300 reads most often Illumina HiSeq MiSeq or NovaSeq protocols.
Paired-end tags are the short sequences at the 5 and 3 ends of a DNA fragment which are unique enough that they exist together only once in a genome therefore making the sequence of the DNA in between them available upon search or upon further sequencing. PacBio SMRT 2017 2018 From PacBio web site. Fast and Accurate Next-Generation Sequencing Results Enabled by Ion Torrent Technology.
Since paired-end reads are more likely to align to a reference the quality of the entire data set. Introduction to Mate Pair Sequencing. Illumina gets sequence data from both strands of input sequence which means it outputs data from both ends of the input and is normally reported two files R1 and R2 often refereed to as mates files R1first mates R2second mates.
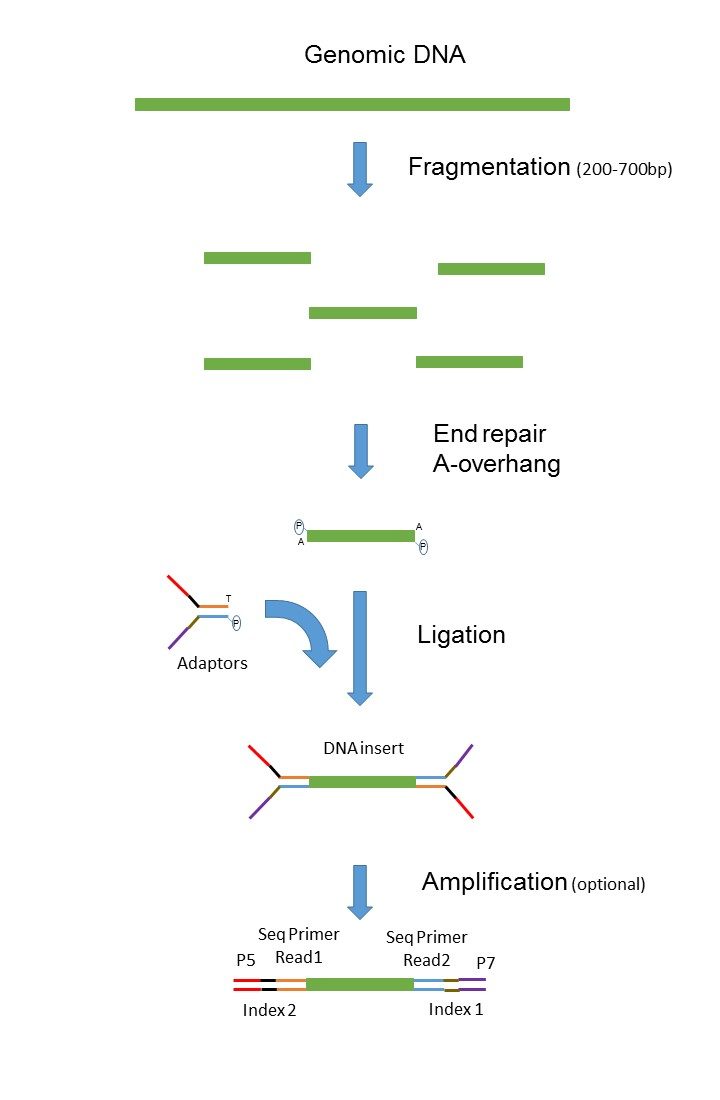
2 For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing. The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases. Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall.
10-20 kb Long-read Sequencing Platform. Illumina Paired End Sequencing. Bases 1-75 forward direction and bases 225-300 reverse direction of the fragment.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Both pairs originate from a single fragment which is sequenced from either end. Mate pair reads Fragment.
Due to the way data is reported in these files special care has to be taken. In DNA sequencing lingo the words paired-end PE and mate-pair MP are frequently used interchangeably. Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. For example if you have a 300bp contiguous fragment the machine will sequence eg. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
1 shows a schematic view of an Illumina paired-end read. Combining data generated from mate pair library sequencing with that from short-insert paired-end reads provides a powerful combination of read lengths for maximal sequencing coverage across the genome. The figure shows the workflow for mate-pair library preparation for Illumina sequencing.
In addition to producing twice the number of reads for the same time and effort in library. Paired-end tags exist in PET libraries with the intervening DNA absent that is a PET represents a larger fragment of. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
Read 1 often called the forward read extends from the Read 1 Adapter in the 5 3 direction towards Read 2 along the forward DNA strand. In single-end reading the sequencer reads a fragment from only one end to the other generating the sequence of base pairs. Since the beginning of 2013 this preparation has been based on Nextera technology.
In the Illumina world the library type are 2. Paired-end library and mate-pair library. While the underlying principles between PE and MP reads have strong similarities there are inherent differences that are crucial to understand.
This is all for conventional paired-end sequencing.

6 Pairwise Strand Of Paired End And Mate Pair Read Libraries Download Scientific Diagram

Ion Torrent Mate Paired Library Preparation Protocol And Sequencing Download Scientific Diagram

Why Do Paired Ends Map To Same Coordinates Of A Scaffold

Principles Of Mate Pair Libraries Construction And The Bioinformatics Download Scientific Diagram

Beyond The Linear Genome Paired End Sequencing As A Biophysical Tool Trends In Cell Biology

Use Of Pairwise Linkage Information For Scaffolding A Paired End Download Scientific Diagram

Mate Pair Made Possible Enseqlopedia

What Is Mate Pair Sequencing For
Paired End Sequencing France Genomique

Construction Of Mate Pair Libraries Download Scientific Diagram
Whole Genome Assembly With Iplant Ppt Video Online Download
Introduction To Genome Assembly Tutorial

What Is Mate Pair Sequencing For

Principles For Construction Of Mate Pair Sequencing Libraries A Download Scientific Diagram
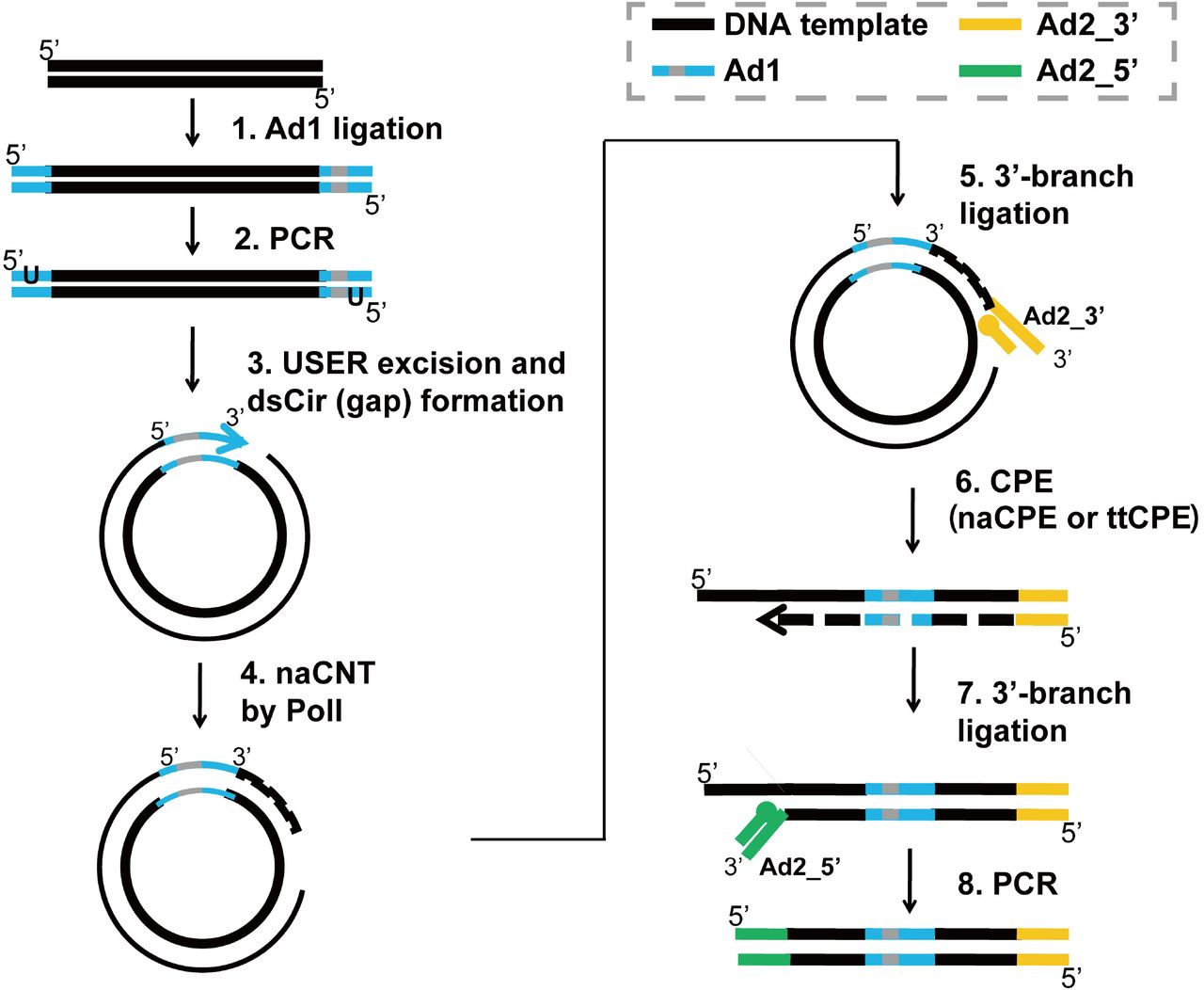
Development Of Coupling Controlled Polymerizations By Adapter Ligation In Mate Pair Sequencing For Detection Of Various Genomic Variants In One Single Assay Biorxiv
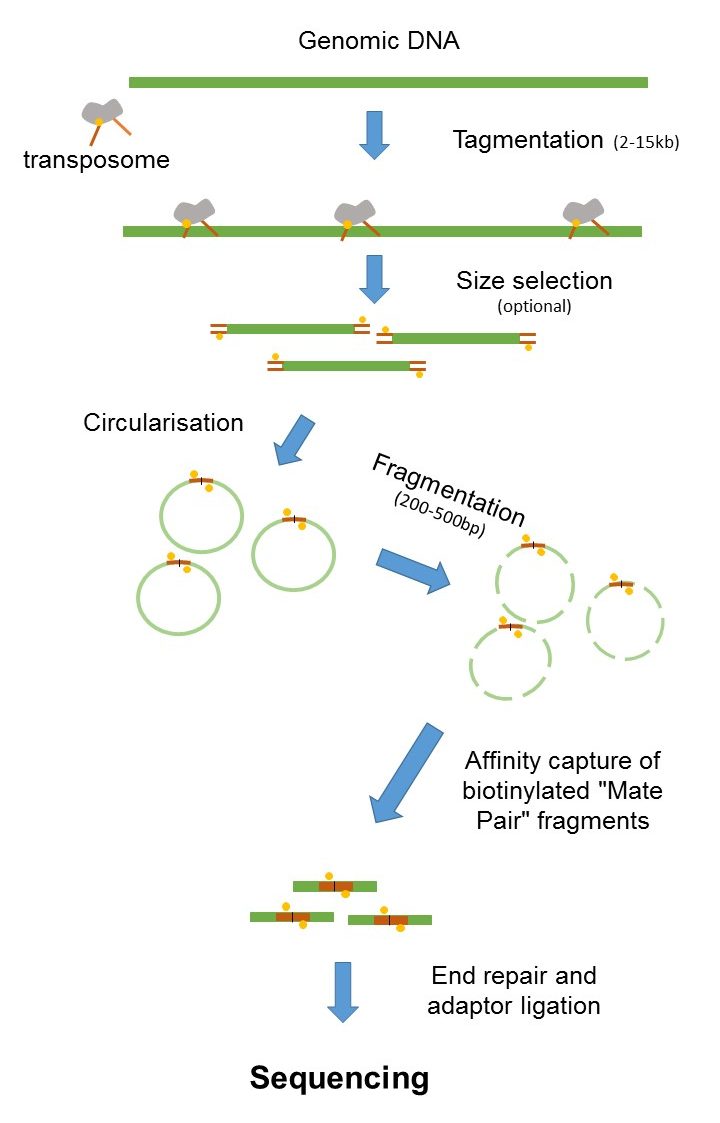
Mate Pair Sequencing France Genomique

Pdf Data Processing Of Nextera Mate Pair Reads On Illumina Sequencing Platforms Semantic Scholar